This category is for questions related to neuron tracing and reconstruction for Neurolucida and Neurolucida 360 software users.
Is it possible, when using a photo taken with several fluorophores, to quantify an intensity of a fluorophore that is not used for modeling?
For example, I am reconstructing neurons using GFP, but also want to quantify the intensity of m-cherry used to detect another protein,
thank you very much in advance!
Find out more on the online help page here: https://www.mbfbioscience.com/help/neurolucida360/Default.htm#cshid=8050.
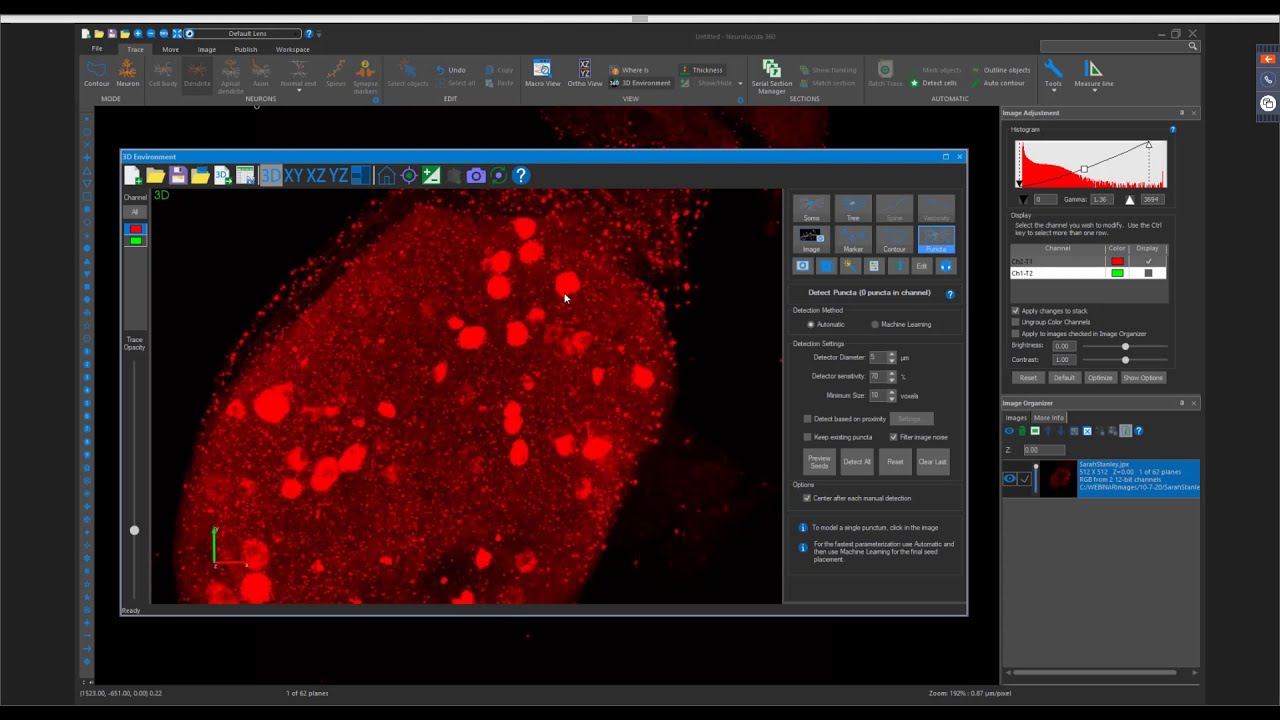
Thank you for the question! You can detect these proteins based on diameter, sensitivity, and voxel size by using Puncta Detection in Neurolucida360. This can give you a spatial understanding of where these proteins are located and the volume/ number of clusters.
There is an upcoming webinar on how to use puncta detection that you can register for here:
https://zoom.us/webinar/register/WN_OTfKFF-vTguM2QpW8BdjLg
And a pre-recorded webinar that is located here:
MBF Bioscience recommends using the puncta detection tool to obtain spatial and volumetric metrics of your labeled protein. Though it is possible to obtain luminance values in our software, we do not advise collecting luminance values to quantify protein concentration because the intensity of a fluorescence signal is determined by many factors including exposure time, tissue processing, light level, bleaching etc. Therefore, intensity of signal does not always linearly correlate with the amount of protein present. You can, however, make qualitative statements about expression between groups using intensity values.
I hope this response is helpful. Please feel free to post any follow-up questions. We are happy to help!