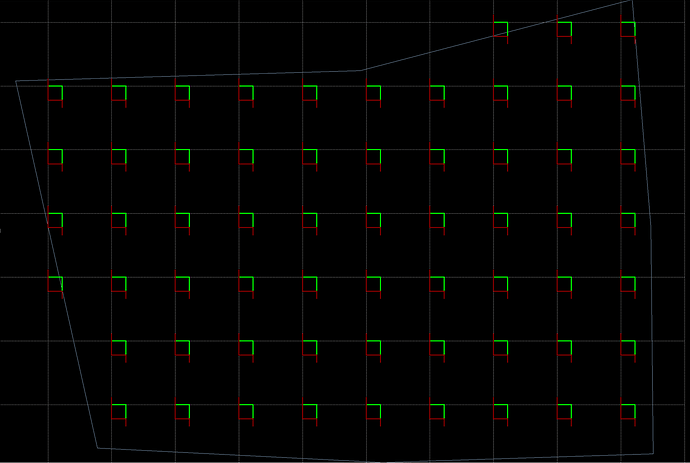
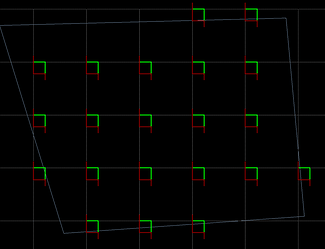
I’m using the optical fractionator and have a few questions:
-
I’m using the same sampling parameters for all of my samples. I want to sample 5% of my region of interest, but the program seems to be making me count more than 5% for about half of my samples (the total number of sampling sites is higher for those animals despite having the same or lower number of sections).
-
I’m counting cells from about 15 animals. Do we need to have the same number of sections for each animal?